亚麻(Linum usitatissimum L.)是一年生的二倍体自花授粉植物,在中国、加拿大和印度广泛种植[1]。它可分为纤维用亚麻和油用亚麻,是纤维和油脂兼用型的作物。油用亚麻简称胡麻,含有多种脂肪酸,其中α-亚麻酸(ALA)含量高达 50%。由于其丰富的营养特性,亚麻已经成为饮食和疾病研究领域日益关注的焦点。近年来,全球范围内极端天气日益频发,加剧了干旱地和盐碱地的形成。因此,要实现农业的可持续发展,培育和筛选耐干旱和耐盐碱的亚麻品种是提高其产量和充分利用逆境土地的有效途径。
虽然亚麻喜凉爽湿润气候,耐寒、耐旱、耐贫瘠,在含盐量0.2%以下的碱性土壤亦能栽培,但是土壤缺水和盐碱化依然会对亚麻的产量、含油量、脂肪酸组分以及纤维的质量产生重大影响[2-6]。亚麻比其他许多油料作物都更耐旱,但它的蒸腾系数很高,因此会损失大量水分[7]。刘莹莹等[8]发现,亚麻在干旱胁迫下,不同生育时期的干物质积累量与产量呈显著正相关关系,影响亚麻籽粒产量的主导因子是千粒重。Quéro等[9]研究发现,b-氨基丁酸(b-aminobutyric acid,BABA)诱导了亚麻的干旱响应过程。BABA会引起亚麻叶片中溶质含量的重组,导致脯氨酸和非结构性碳水化合物的积累,使亚麻更耐干旱。土壤盐碱化会导致亚麻发芽和出苗延迟、幼苗存活率低、生长发育不稳定以及减产。通过对亚麻的种质资源进行盐碱胁迫,根据生物量、发芽率、幼苗性状和产量筛选得到一批耐盐碱品系[10-14]。为了解析亚麻耐盐性的遗传基础,Yu等[15]利用深度测序技术分析了3种胁迫条件下样本中的小RNA和降解物。研究发现,miR398a和miR530这2个miRNA与亚麻的盐胁迫耐受性有关。将亚麻在碱-盐胁迫、中性盐胁迫和碱性胁迫下处理[16]发现,在中性盐胁迫下,碳水化合物代谢受到影响;在碱-盐胁迫下,光合作用和对生物刺激的反应受到严重影响。碱-盐胁迫下调控的差异表达基因多于碱性盐胁迫或中性盐胁迫下调控的差异表达基因,表明有更多的基因参与调控碱-盐胁迫途径。
植物中存在一个复杂而精细的信号调控网络应对干旱、高盐和低温等环境胁迫,通过调控不同信号网络之间的转录因子和信号转导的蛋白酶类参与胁迫应答。不同的环境胁迫下,特定的转录因子通过与启动子顺式作用元件相结合激活或抑制下游靶基因的表达,在信号转导过程中发挥着关键作用。目前已鉴定出多个参与非生物胁迫调控的转录因子,如NCED、LEA、DREB、MYB/C、ABREs和AP2/ERF转录因子[17-18]。其中,AP2/ERF类转录因子是调控植物生长发育和胁迫响应的关键调节因子[5,19-22],与植物生长发育[23-24]、生长激素[25-27]、低温[28]、干旱[29-30]以及高盐[31-34]的胁迫应答等密切相关。
AP2/ERF是植物转录因子最大的家族之一[35],已从多种植物中鉴定出AP2/ERF转录因子[36],如拟南芥、水稻[37]、小麦[38]、大豆[39]和葡萄[40]。AP2/ERF包含1个或2个非常保守的AP2/ERFDNA结合域(DNA-binding domain),至少包含一个高度保守的AP2/ERFDNA结合结构域。大约由60~70个氨基酸残基组成,可直接与下游靶基因启动子上的GCC盒和/或脱水反应元件(DRE)/C-重复元件(CRT)等顺式作用元件相互作用[36]。根据AP2/ERFDNA结合结构域的数量和相似性,可分为5个亚家族:AP2(APETALA2)、RAV(与ABI3/VP1有关)、DREB(脱水反应元件结合蛋白)、ERF(乙烯反应因子)和其他因子[41]。其中AP2家族蛋白拥有2个重复的AP2结构域[42]。近年来,很多研究表明过表达AP2/ERF类转录因子在植物的耐旱和耐盐性中具有重要作用[43-45]。水稻(Oryzasativa.L.)OsERF83是一个定位于细胞核的转录因子,在干旱和脱落酸(ABA)胁迫时被诱导,过表达OsERF83可以显著提高转基因水稻的抗旱性,并增强了光化学效率[46]。ZmERF21(ZeamaysL.)在玉米中过表达显著增加了干旱条件下的叶绿素含量和抗氧化酶的活性,ZmERF21可能通过与潜在靶基因的启动子结合,直接调节与激素(乙烯、脱落酸)和Ca信号相关的基因以及其他应激响应基因的表达,从而增强玉米幼苗的耐旱性[47]。陆地棉GhERF13.12(Gossypiumhirsutum)在拟南芥中异源表达可以增强转基因植物的耐盐性,同时上调ABA信号传递、脯氨酸生物合成和ROS清除途径的相关基因的表达。此外,沉默GhERF13.12导致棉花对盐胁迫的耐受性降低[48]。从耐盐甘薯系(Ipomoea batatas (L.) Lam.)ND98中克隆了一个AP2/ERF基因IbRAP2-12,在拟南芥中异源表达IbRAP2-12可以增强盐胁迫和干旱胁迫的耐受性。在盐和干旱胁迫下,参与ABA信号、JA信号、脯氨酸生物合成和活性氧(ROS)清除途径中的基因在IbRAP2-12过表达株系中被显著上调[49]。
WRINKLED1(WRI1)是AP2/ERF类转录因子家族成员,调节碳在糖酵解和脂肪酸生物合成途径之间的分配[50],之前有很多研究表明AtWRI1或WRI1的同源基因可以显著提高转基因植物种子的含油量[51-55],但是在植物耐逆方面的功能作用还鲜有报道。前期我们从亚麻中克隆了一个WRINKLED1的同源基因LuWRI1a,蛋白序列分析LuWRI1a包含2个AP2的DNA结合域,属于AP2转录因子家族[56]。通过分析LuWRI1a的顺式作用元件,发现含有响应光、干旱、低温和激素等多个非生物胁迫应激元件。本研究通过对LuWRI1a过表达转基因植株进行NaCl盐胁迫和PEG-6000模拟干旱胁迫处理,测定各项生长指标和生理指标,分析转录因子LuWRI1a在亚麻盐胁迫和干旱胁迫应答反应中的功能,有助于揭示AP2/ERF类转录因子在逆境环境下的表达调控机制,为亚麻耐拟品种的改良提供新的候选基因。
1 材料与方法
1.1 试验材料
供试材料为亚麻(Linum usitatissimum)栽培品种陇亚10号和LuWRI1a过表达T3代纯合转基因株系LuWRI1a-OX-X,种子由甘肃省农业科学院作物研究所亚麻研究室提供。
1.2 LuWRI1a过表达亚麻株系的获得
以非转基因“陇亚10号”(野生型)为阴性对照。通过农杆菌介导法转化陇亚10号,将包含重组质pE101-BASTA-LuWRI1a的农杆菌GV3101侵染亚麻下胚轴,转化方法参照文献[57]。先后在不定芽诱导培养基和生根培养基中进行培养[58],每2周更换1次培养基,直至获得完整植株,收获T0代种子。将T0代种子播种在含有10mgmL-1Basta(Sigma Aldrich)的1/2MS培养基上,取抗性苗叶片用CTAB法提取DNA,用表达载体上的bar基因序列设计特异引物进行PCR检测,收获T1代种子。通过Basta筛选和PCR检测鉴定阳性植株,直至获得T3代纯合株系,用于后续试验。
1.3 LuWRI1a 家族成员启动子分析
从亚麻基因组序列中提取 LuWRI1a 基因起始位置上游 2000 bp 的序列,在PlantCARE数据库(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)进行顺式作用元件分析。
1.4 试验处理
将进口的丹麦品质草炭土和蛭石按3:1的体积混合均匀,取适量混合营养土装入小花盆中,盆高18cm,直径15cm。选取籽粒饱满、大小均匀的亚麻种子约15粒,播种到花盆内,室温放置。用1/2Hogland营养液培养1个月,每隔3d浇灌100mL次-1。光照强度为125μmolm-2s-1,光周期为16h/8h(光照/黑暗),将培养4周的幼苗进行胁迫处理。试验设置4个处理,3次重复。挑选长势相同的幼苗进行如下处理:①盐胁迫处理:用1/2Hoagland营养液配制200mmolL-1NaCl营养液,每隔3d浇灌100mL;②PEG-6000模拟干旱处理:用1/2Hoagland营养液配制25%PEG营养液,每隔3d浇灌100mL;对照组每隔3d浇灌100mL1/2Hogland营养液。胁迫处理2周后,观察表型并测定相关生理指标。每个株系选取9株进行株高、根长、侧根数和叶片数的测定。此外,每组不同处理的材料随机挑选3株亚麻,剪取根部组织和地上组织,每个株系取3个重复。液氮速冻后于-80℃冰箱储存。
1.5 测定与方法
胁迫处理2周后,使用苏州科铭生物试剂盒测定超氧化歧化酶(Super oxide dismutase,SOD)活性、过氧化氢酶(Catalase,CAT)活性、丙二醛(Malondialdehyde,MDA)含量以及抗坏血酸过氧化物酶(Ascorbate peroxidase,APX)。。
1.6 超声微波协同提取法引物合成及基因表达分析
根据本研究室公布的亚麻基因组序列(NCBI,http://www.ncbi.nlm.nih.gov/,登录号为QMEI02000000)CDS序列以及蛋白质序列(https://doi.org/10.6084/m9.figshare.13614311.v3),利用Primer5.0设计鉴定阳性植株的PCR引物和荧光定量引物。利用qRT-PCR方法检测WRI1以及耐逆相关的4个关键基因LuAREB(L.us.o.m.scaffold233.70)、LuDREB(L.us.o.m.scaffold284.37)、LuLEA(L.us.o.m.scaffold47.25)、LuNCED(L.us.o.m.scaffold66.130)在对照和转基因株系中的表达模式[59]。以甘油醛-3-磷酸(GAPDH,Glyceraldehyde3-phosphatedehydrogenase)为内参基因。由上海生物化工公司合成所有引物(表1)。实时荧光定量PCR方法参照文献[59]。
表1 PCR引物
1.7 数据分析
用Microsoft Excel 2010程序进行原始数据的整理,用SPSS 20.0软件进行数据整理及统计分析。利用GraphPad Prism8. 0软件作图。
2 结果与分析
2.1 LuWRI1a 启动子顺式作用元件分析
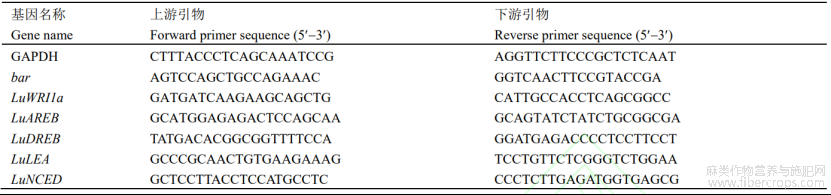
应用PlantCARE软件对基因启动子区进行分析(图1)发现,pLuWRI1a含有丰富的顺式作用元件,除了含有TATA-BOX、CAAT-box等核心功能响应元件外,还包含多个其他类型的顺式作用元件。根据功能注释可分为与光响应相关的顺式作用元件(ACE、AE-box、GT1-motif、Box4和Gap-box),与激素响应有关的顺式作用元件(TGACG-motif、CGTCA-motif和TCA-element),与发育相关的元件(MBSI、O2-site),以及与非生物胁迫响应的相关元件(MBS、LTR、MYB、MYC和ARE),结果见表2。因此推测,LuWRI1a可能参与盐胁迫、干旱等非生物逆境胁迫响应等表达调控途径。
图1 亚麻LuWRI1a启动子顺式作用元件
表2 LuWRI1a 启动子顺式作用元件的推测功能
2.2 LuWRI1a转基因亚麻的获得和分子鉴定
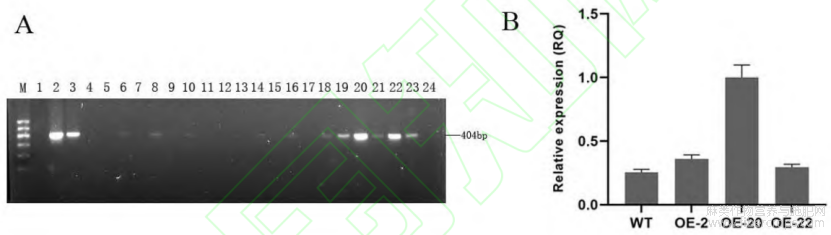
通过农杆菌介导法将过表达载体P101--BASTA-LuWRI1a转化亚麻栽培品种陇亚10号的下胚轴,获得转化苗(图2),使用载体上的bar基因特异引物对转化苗进行PCR鉴定(图3-A),获得7个转基因株系。根据qRT-PCR的检测结果,选择表达量较高的3个不同的转基因株系OE-2、OE-20和OE-22进行下一步试验(图3-B)。
图2 转基因亚麻的获得
图3 转基因亚麻的分子鉴定
2.3 转基因植株的耐逆性分析
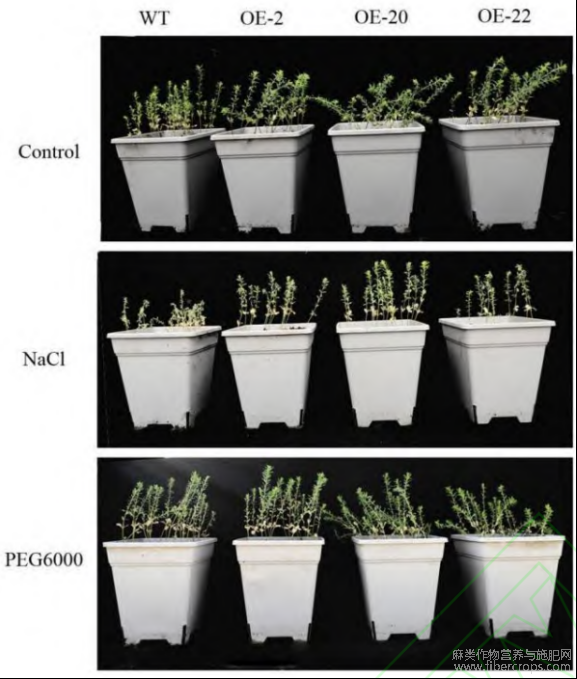
为进一步研究LuWRI1a的耐旱性和耐盐性,分别用25%的PEG-6000和200mmolL-1的NaCl对野生型亚麻(陇亚10号)和转基因亚麻进行2周胁迫处理。结果显示,转基因株系的株高、主根长、侧根数和叶片数在正常培养条件下与野生型相比均无明显差异。在200mmolL-1NaCl胁迫下,野生型亚麻株高较低,叶片发黄,严重干枯,萎蔫程度较转基因株系严重,这表明过表达LuWRI1a基因亚麻的耐盐能力比野生型亚麻更强。PEG-6000胁迫处理后,野生型植株和转基因株系均未出现萎蔫,转基因株系叶片部分失绿,表型无明显差异(图4)。
图4 胁迫处理后野生型植株和转基因植株的表型
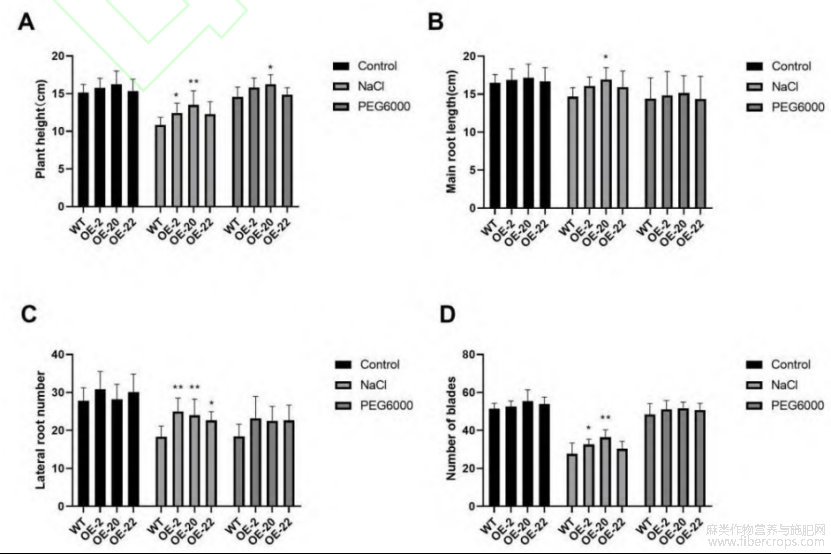
转基因植株在盐胁迫处理后,相对株高、侧根数及叶片数与野生型相比差异极显著;在干旱胁迫处理后,各指标与野生型相比差异不显著。表明过表达LuWRI1a的转基因株系对盐胁迫的耐受性更高(图5)。盐胁迫处理2周后,野生型植株的生长受到了抑制,侧根数目明显减少,转基因植株的平均侧根数相比对照分别提高36.36%、30.91%和23.63%。平均根长分别提高9.46%、15.44%和8.63%,平均叶片数分别提高18.07%、31.73%和9.64%;干旱胁迫处理2周后,转基因植株与对照相比无显著差异,平均主根长分别提高3.08%、5.17%和负0.23%,平均侧根数分别提高25.9%、22.29%和22.89%,平均叶片数分别提高5.5%、6.88%和4.59%。
图5 盐胁迫和干旱胁迫下转基因植株性状分析
2.4 非生物胁迫对转基因植株抗氧化物质酶活性和MDA含量的影响
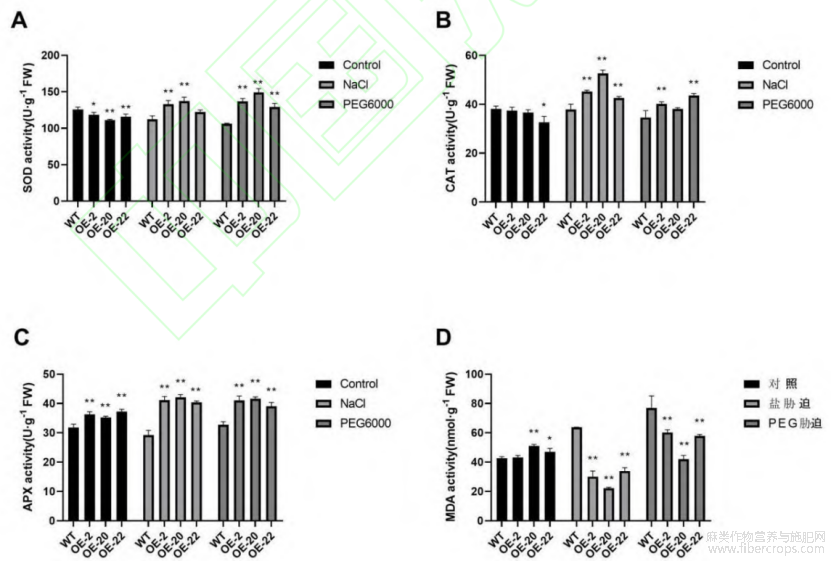
超氧化歧化酶(SOD)活性、过氧化氢酶(CAT)活性和抗坏血酸过氧化物酶(APX)活性以及丙二醛(MDA)的含量是植物在耐逆条件下的重要生理指标。正常培养后,转基因植株的SOD活性比野生型植株的低,APX 活性比野生型植株的高,CAT 活性和 MDA 含量并无明显差异。盐胁迫和干旱胁迫后,3 种抗氧化酶的活性与对照相比极显著升高,而 MDA 含量极显著降低。
盐胁迫处理后,野生型亚麻的SOD活性为112.36Ug-1,转基因亚麻的SOD酶活性分别为133.02、137.21和122.25Ug-1,比野生型提高0.18%、0.22%和0.09%(图6-A);野生型亚麻的CAT酶活性为37.85Ug-1,转基因亚麻的CAT酶活性分别为45.19、52.62和42.63Ug-1,比野生型提高0.19%、0.39%和0.13%(图6-B);野生型亚麻的APX酶活性为29.24Ug-1,转基因亚麻的APX酶活性分别为41.19、42.13和40.41Ug-1,比野生型提高0.41%、0.44%和0.38%(图6-C);野生型亚麻的MDA含量为63.75nmol g-1,转基因亚麻的MDA含量分别为30.11、22.22和33.93nmol g-1,比野生型亚麻降低0.53%、0.65%和0.47%(图6-D)。
干旱胁迫处理后,野生型亚麻的SOD活性为106.27Ug-1,转基因亚麻的SOD酶活性分别为136.73、149.00和129.02Ug-1,比野生型提高0.28%、0.40%和0.21%(图6-A);野生型亚麻的CAT酶活性为34.63Ug-1,转基因亚麻的CAT酶活性分别为40.18、38.11和43.63Ug-1,比野生型提高0.16%、0.10%和0.26%(图6-B);野生型亚麻的APX酶活性为32.74Ug-1,转基因亚麻的APX酶活性分别为41.06、41.61和39.06Ug-1,比野生型提高0.25%、0.27%和0.19%(图6-C);野生型亚麻的MDA含量为76.95nmol g-1,转基因亚麻的MDA含量分别为60.27、42.08和57.94nmol g-1,比野生型亚麻降低0.22%、0.45%和0.25%(图6-D)。
图6 过表达LuWRI1a亚麻的酶活性测定
2.5 逆境胁迫响应基因在转基因亚麻中的表达分析
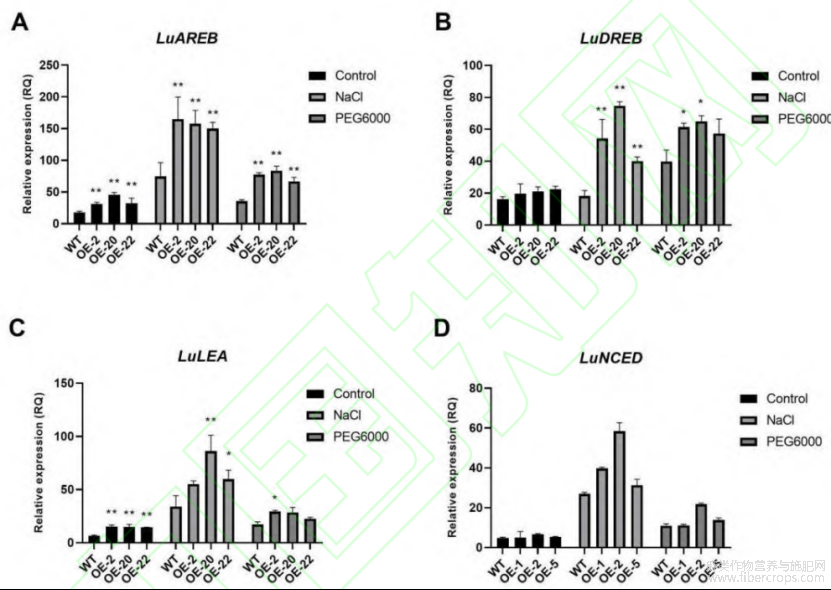
为研究 LuWRI1a 在逆境胁迫响应中可能的分子机制,本研究分析了非生物胁迫响应途径中的 4 个关键转录调控因子,AREB2( ABA-responsive element binding)、DREB2(dehydration-responsive element binding)、LEA(late embryogenesis-abundant protein)和NCED(9-cis-epoxycarotenoid dioxygenase)在野生型亚麻和转基因亚麻中的表达水平[60]。通过实时荧光定量(qRT-PCR)分析表明,在盐胁迫和干旱胁迫的处理下,这 4 个逆境胁迫相关基因均上调表达(图 7)。LuAREB2 在正常和逆境胁迫条件下,转基因植株中的表达量与野生型相比差异极显著(图 7-A)。LuDREB2 在正常条件下,野生型和转基因植株中的表达量没有明显差异,在干旱胁迫处理后较对照显著上调,而在盐胁迫处理后极显著上调(图 7-B)。LuLEA 在正常培养和盐胁迫处理后,转基因植株中的表达量均极显著高于野生型,但在干旱胁迫处理后,株系间差异不显著(图 7-C)。正常条件下和干旱胁迫处理后,LuNCED 较对照无明显差异,在盐胁迫处理后,基因的表达量升高,但株系间差异不显著(图 7-D)。表明,在逆境胁迫下,尤其在盐胁迫下,转基因植株中 LuWRI1a 通过上调 LuAREB、LuDREB、LuLEA 和 LuNCED 等逆境胁迫响应基因的表达,来参与调控亚麻转基因植株的耐逆性。
图7 胁迫处理下转基因亚麻逆境胁迫相关基因的表达分析
3 讨论
干旱、高盐、极端天气、洪水和病虫害等各种生物和非生物胁迫对植物的生长和发育产生负面影响。目前,非生物胁迫已被认为是造成全世界作物损失的主要因素之一,并导致大多数主要农作物产量下降高达50%[61]。在非生物胁迫下,植物中的转录因子与胁迫应答基因启动子的顺式作用元件结合,通过在转录水平上激活或抑制应答基因,调节逆境胁迫给植物带来的损伤[5,17]。近年来,AP2/ERF转录因子家族备受关注。研究表明,AP2/ERF转录因子可调控植物发育和胁迫响应的多种过程,如生殖发育、细胞增殖、非生物和生物胁迫响应以及植物激素响应。因此,阐明AP2/ERF在胁迫信号中的调控机制非常重要,可以通过调控AP2/ERF来提高作物的抗逆性。
WRI1属于AP2/ERF家族转录因子,前期WRI1的研究大多集中在种子含油量方面。例如,在拟南芥中过表达AtWRI1,种子含油量增加大约10%~20%[62]。在玉米中过表达ZmWRI1,种子含油量提高了大约31%[63],在拟南芥和油菜中过表达BnWRI1,种子含油量分别增加了约20%和10%[64]。近年来,有一些研究表明WRI还有可能参与植物非生物胁迫信号途径。大豆GmWRI1a(Glycinemax)启动子存在乙烯、茉莉酸、赤霉素3种激素响应元件,受这3种激素胁迫诱导[65-66],并受ABA、NaCl和糖信号等调控[67]。花生AhWRI1-1基因对3种非生物胁迫均有响应。低温胁迫下表达量明显上调,在干旱胁迫下明显下调。AhWRI1-2基因在高盐、低温、干旱非生物胁迫下表达量均有上调。AhWRI1-1和AhWRI1-2这2个基因可能参与了花生对高盐、低温、干旱的抗性调节[68]。
生长指标方面,盐胁迫后野生型亚麻萎蔫程度较转基因株系严重,干旱胁迫后表型差异不明显;转基因植株的相对株高、主根长度、侧根数目及叶片数在盐胁迫和干旱胁迫处理后均升高,尤其在盐胁迫处理后,转基因植株各指标均明显高于对照,表明过表达LuWRI1a的转基因株系对盐胁迫的耐受性更高。这个结果说明生长抑制是亚麻对盐胁迫和干旱胁迫的主要反应之一,LuWRI1a能抵抗盐胁迫和干旱胁迫对亚麻生长的抑制。
活性氧(reactive oxygen species,ROS)是指在生物体内与氧代谢有关的、含氧自由基和易形成自由基的过氧化物的总称。当植物在逆境胁迫时,会诱导植物细胞中活性氧的积累,ROS的过度积累导致会导致细胞产生氧化应激损伤[69],从而进一步产生有毒物质[70]。活性氧清除酶系统是清除过量的活性氧物质,维持植物体内动态平衡最有效的机制[71],一般包括超氧化物歧化酶(SOD)、抗坏血酸过氧化物酶(APX)、过氧化氢酶(CAT),植物通过这些酶类抗氧化剂维持细胞的正常发育。通过测定抗氧化物质酶活性,发现胁迫处理后,转基因株系的3种抗氧化酶APX、SOD和CAT的活性比野生型均显著性升高。丙二醛(MDA)能加剧膜脂损伤,它的含量代表膜脂过氧化的程度[72]。盐胁迫和干旱胁迫处理后,野生型株系的MDA含量均比处理前升高,而转基因株系的MDA含量均显著低于野生型。这些结果表明过表达LuWRI1a能够增强在盐胁迫和干旱胁迫下亚麻的抗氧化能力,通过减轻对细胞膜造成的氧化损伤,增强耐逆性。
实时荧光定量(qRT-PCR)分析表明,在盐胁迫和干旱胁迫的处理下,4个逆境胁迫相关基因LuAREB2、LuDREB2、LuLEA和LuNCED均上调表达,说明LuWRI1a可能是通过激活LuAREB2、LuDREB2、LuLEA和LuNCED等逆境胁迫响应基因的表达增强转基因植株的耐逆性。
本研究结果表明,亚麻转录因子LuWRI1a启动子区含有多个参与非生物胁迫应答的响应元件。进一步挖掘LuWRI1a在逆境胁迫下的的功能,发现LuWRI1a通过抵抗盐胁迫和干旱胁迫对亚麻生长的抑制,增强活性氧清除能力、减轻膜脂的氧化损伤,激活逆境胁迫响应基因的表达等途径,增强了亚麻的耐逆性。
4 结论
综上所述,LuWRI1a可能是一个多功能基因,它不仅参与脂肪酸合成代谢途径,还有可能参与植物非生物胁迫信号途径。本研究为亚麻耐逆品种改良提供了新的基因资源,为培育亚麻耐逆新品系提供理论依据。
[1] Huis R, Hawkins S, Neutelings G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.).BMC Plant Biol, 2010, 10: 14.
[2] Zheng J, Cui B, Yan Y H, Gao B, Wu Y F, Wang H D, Wang P, Xu B Q, Zhao Z, Cao Y, Zhang Y P. Agronomic cultivation measures on productivity of oilseed flax: A review. Oil Crop Sci, 2022, 7: 53–62.
[3] Zare S, Mirlohi A, Saeidi G, Ataii E. Water stress intensified the relation of seed color with lignan content and seed yield components in flax (Linum usitatissimum L.). Sci Rep, 2021, 11: 23958.
[4] Fila G, Bagatta M, Maestrini C, Potenza E, Matteo R. Linseed as a dual-purpose crop: evaluation of cultivar suitability and analysis of yield determinants. J Agric Sci, 2018, 156: 1−15.
[5] Zhang J, Liao J, Ling Q, Xi Y, Qian Y. Genome-wide identification and expression profiling analysis of maize AP2/ERF superfamily genes reveal essential roles in abiotic stress tolerance. BMC Genomics, 2022, 23: 125.
[6] Yadav B, Kaur V, Narayan O P, Yadav S K, Kumar A, Wankhede D P. Integrated omics approaches for flax improvement under abiotic and biotic stress: Current status and future prospects. Front Plant Sci, 2022, 13: 931275.
[7] Paliwal S, Tripathi M K, Tiwari S, Tripathi N, Payasi D K, Tiwari P N, Singh K, Yadav R K, Asati R, Chauhan S. Molecular advances to combat different biotic and abiotic stresses in Linseed (Linum usitatissimum L.): a comprehensive review. Genes (Basel), 2023, 14: 1461.
[8] 刘莹莹, 李玥, 吴兵. 胡麻籽粒产量形成对干旱胁迫的响应及其模拟模型研究. 作物研究, 2023, 37(1): 14−21.Liu Y Y, Li Y, Wu B. Response of kernel yield formation to drought stress and its simulation modeling in flaxseed. Crop Res, 2023. 37(1): 14−21(in Chinese with English abstract).
[9] Kariuki L W, Masinde P, Githiri S, Onyango A N. Effect of water stress on growth of three linseed ( Linum usitatissimum L.) varieties.Springerplus, 2016, 5: 1−16.
[10] EL-Afry M M, EL-Okkiah S A F, EL-Kady E-S A F, EL-Yamanee G S A. Exogenous application of ascorbic acid for alleviation the adverse effects of salinity stress in flax (Linum usitatissimum L.). Middle East J Agric Res, 2018, 7: 716−739.
[11] Nasri N, Maatallah S, Saidi I, Lachal M. Influence of salinity on germination, seedling growth, ion content and acid phosphatase activities ofLinum usitatissimum L. J Anim Plant Sci, 2017, 27: 517−521.
[12] Datir S. Salt-induced physiological and biochemical changes in two varieties of Linum usitatissimum L. Int J Curr Microbiol Appl Sci, 2015, 4: 296−304.
[13] Demir Kaya M, Day S, Cikili Y, Arslan N. Classification of some linseed (Linum usitatissimum L.) genotypes for salinity tolerance using germination, seedling growth, and ion content. Chilean J Agric Res, 2012, 72: 27−32.
[14] 于莹, 陈宏宇, 程莉莉, 赵东升, 袁红梅, 吴广文, 关凤芝. 亚麻 MAPK 基因克隆及盐碱胁迫下的表达分析. 东北农业大学学报, 2015,46(3): 8.Yu Y, Chen H Y, Cheng L L, Zhao D S, Yuan H M, Wu G W, Guan F Z. Flax MAPK gene cloning and expression analysis under saline and alkaline stress. J Northeast Agric Univ, 2015, 46(3): 8 (in Chinese with English abstract).
[15] Yu Y, Chen H, Yang Y Y, Lou D, Liang C, Yuan H, Wu G W, Xu C. Identification and characterization of differentially expressed microRNAs and target gene related to flax stem development. J Nat Fibers, 2021, 19: 5974−5990.
[16] Guo R, Zhou J, Ren G X, Hao W. Physiological responses of linseed seedlings to iso osmotic polyethylene glycol, salt, and alkali stresses. Agron J, 2013, 105: 764.
[17] 郭晋艳, 郑晓瑜, 邹翠霞, 李秋莉. 植物非生物胁迫诱导启动子顺式元件及转录因子研究进展. 生物技术通报, 2011, 23(4): 16−20.
Guo J Y, Zheng X Y, Zou C X, Li Q L. Progress of abiotic stress-induced promoter cis-elements and transcription factors in plants. Biotechnol Bull, 2011, 23(4): 16−20 (in Chinese with English abstract).
[18] J L Riechmann , J Heard, G Martin, L Reuber, C Jiang, J Keddie, L Adam, O Pineda, O J Ratcliffe, R R Samaha, R Creelman, M Pilgrim, P Broun, J Z Zhang, D Ghandehari, B K Sherman, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science, 2000, 290: 2105−2110.
[19] Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, Ohme-Takagi M. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol, 2011, 21: 508−514.
[20] Jofuku K D, den Boer B G, Van Montagu M, Okamuro J K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell, 1994, 6: 1211−1225.
[21] Jaglo-Ottosen K R, Gilmour S J, Zarka D G, Schabenberger O, Thomashow M F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science, 1998, 280: 104−116.
[22] Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Novartis Found Symp, 2001, 290: 2105−2110.
[23] Chandler J W, Cole M, Flier A, Grewe B, Werr W. The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development, 2007, 134: 1653−1662.
[24] Aoyama T, Hiwatashi Y, Shigyo M, Kofuji R, Kubo M, Ito M, Hasebe M. AP2-type transcription factors determine stem cell identity in the moss Physcomitrella patens. Development, 2012, 139: 3120−3129.
[25] De Boer K, Tilleman S, Pauwels L, Vanden Bossche R, De Sutter V, Vanderhaeghen R, Hilson P, Hamill J D, Goossens A. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant Journal, 2011, 66: 1053−1065.
[26] Finkelstein R R, Wang M L, Lynch T J, Rao S, Goodman H M. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell, 1998, 10: 1043−1054.
[27] Lorenzo O, Piqueras R, Sánchez-Serrano J J, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell, 2003, 15: 165−178.
[28] Cook D, Fowler S, Fiehn O, Thomashow M F. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA, 2004, 101: 15243−15258.
[29] Cheng M C, Hsieh E J, Chen J H, Chen H Y, Lin T P. Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol, 2012, 158: 363−375.
[30] Oh S J, Kim Y S, Kwon C W, Park H K, Jeong J S, Kim J K. Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol, 2009, 150: 1368−1379.
[31] Chen X, Guo Z. Tobacco OPBP1 enhances salt tolerance and disease resistance of transgenic rice. Int J Mol Sci, 2008, 9: 2601−2613.
[32] Seo Y J, Park J B, Cho Y J, Jung C, Seo H S, Park S K, Nahm B H, Song J T. Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol Cells, 2010, 30: 271−277.
[33] Song C P, Galbraith D W. AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol Biol, 2006, 60: 241−257.
[34] Schmidt R, Mieulet D, Hubberten H M, Obata T, Hoefgen R, Fernie A R, Fisahn J, San Segundo B, Guiderdoni E, Schippers J H, Mueller-Roeber B. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell, 2013, 25: 2115−2131.
[35] Wessler S R. Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci, 2005, 10: 54−66.
[36] Xu Z S, Cheng M, Li L C, Ma Y Z. Functions and application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol, 2011, 53: 570−585.
[37] 靳鹏, 黄立钰, 王迪, 吴慧敏, 朱苓华, 傅彬英. 水稻 AP2/EREBP 转录因子响应非生物胁迫的表达谱分析. 中国农业科学, 2009, 42:3765−3773.Jin P, Huang L Y, Wang D, Wu H M, Zhu L H, Fu B Y. Expression profiling of rice AP2/EREBP transcription factors in response to abiotic stress.Scientia Aguicultura Sinica, 2009. 42: 3765−3773 (in Chinese with English abstract).
[38] Xu Z S, Ni Z Y, Liu L, Nie L N, Li L C, Chen M, Ma Y Z. Characterization of the TaAIDFa gene encoding a CRT/DRE-binding factor responsive to drought, high-salt, and cold stress in wheat. Mol Genet Genomics, 2008, 280: 497−508.
[39] Zhang G, Chen M, Chen X, Xu Z, Guan S, Li L C, Li A, Guo J, Mao L, Ma Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J Exp Bot, 2008, 59: 4095−4107.
[40] Licausi F, Giorgi F M, Zenoni S, Osti F, Pezzotti M, Perata P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics, 2010, 11: 1−15.
[41] Sakuma Y, Liu Q, Dubouzet J G, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun, 2002, 290: 998−1009.
[42] Kagaya Y, Ohmiya K, Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res, 1999, 27: 470−478.
[43] Gao S, Zhang H, Tian Y, Li F, Zhang Z, Lu X, Chen X, Huang R. Expression of TERF1 in rice regulates expression of stress-responsive genes and enhances tolerance to drought and high-salinity. Plant Cell Rep, 2008, 27: 1787−1795.
[44] Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot, 2009, 60: 3781−3796.
[45] Zhang H, Liu W, Wan L, Li F, Dai L, Li D, Zhang Z, Huang R. Functional analyses of ethylene response factor JERF3 with the aim of improving tolerance to drought and osmotic stress in transgenic rice. Transgenic Res, 2010, 19: 809−818.
[46] Eun J S, Woon B S, Hwan K S, Sung S J, Bin Y H, Shic K Y, Kon K J. Overexpression of OsERF83, a Vascular Tissue-Specific Transcription Factor Gene, Confers Drought Tolerance in Rice. Multidiscipl Digital Publish Instit, 2021, 22: 1−20.
[47] Wang Z, Zhao X, Ren Z, Abou-Elwafa S F, Pu X, Zhu Y, Dou D, Su H, Cheng H, Liu Z, Chen Y, Wang E, Shao R, Ku L. ZmERF21 directly regulates hormone signaling and stress-responsive gene expression to influence drought tolerance in maize seedlings. Plant Cell Environ, 2022, 45: 312−328.
[48] Lu L L, Qanmber G , Li J, Pu M L, Chen G Q, Li S D, Liu L, Qin W Q, Ma S Y, Wang Y, Chen Q J, Liu Z. Identification and characterization of the ERF subfamily B3 group revealed GhERF13.12 improves salt tolerance in upland cotton. Frontiers in Plant Science, 2021, 12: 1−15.
[49] Li Y, Zhang H, Zhang Q, Liu Q, Zhai H, Zhao N, He S. An AP2/ERF gene, IbRAP2-12, from sweetpotato is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Sci, 2019, 281: 19−30.
[50] Fei W, Yang S, Hu J, Yang F, Qu G, Peng D, Zhou B. Research advances of WRINKLED1 (WRI1) in plants. Funct Plant Biol, 2020, 47: 185−194.
[51] Cernac A, Benning C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis inArabidopsis. Plant J, 2004, 40: 575−585.
[52] Liu J, Hua W, Zhan G, Wei F, Wang X, Liu G, Wang H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Biochem, 2010, 48: 9−15.
[53] Yang Y, Munz J, Cass C, Zienkiewicz A, Kong Q, Ma W, Sedbrook J, Benning C. Ectopic expression of WRINKLED1 affects fatty acid homeostasis in brachypodium distachyon vegetative tissues. Plant Physiol, 2015, 169: 1836−1847.
[54] Sun R, Ye R, Gao L, Zhang L, Wang R, Mao T, Zheng Y, Li D, Lin Y. Characterization and ectopic expression of coWRI1, an AP2/EREBP domain-containing transcription factor from Coconut (Cocos nucifera L.) endosperm, changes the seeds oil content in transgenic Arabidopsis thaliana and rice (Oryza sativa L.). Front Plant Sci, 2017, 8: 63.
[55] Ye J, Wang C, Sun Y, Qu J, Mao H, Chua N-H. Overexpression of a transcription factor increases lipid content in a woody perennial Jatropha curcas. Front Plant Sci, 2018, 9: 1479.
[56] Li W, Wang L, Qi Y, Xie Y, Zhao W, Dang Z, Zhang J. Overexpression of WRINKLED1 improves the weight and oil content in seeds of flax (Linum usitatissimum L.). Front Plant Sci, 2022, 13: 1003758.
[57] 陈芳. 亚麻FAD3 基因的克隆及载体构建与遗传转化. 甘肃省农业大学硕士学位论文, 甘肃兰州, 2014.Chen F. Cloning and Vector Construction and Genetic Transformation of Flax FAD3 Gene. MS Thesis of Gansu Agricultural University, Lanzhou, Gansu, China, 2014 (in Chinese with English abstract).
[58] 陈芳, 党占海, 张建平, 李闻娟, 郝荣楷, 张琼, 张瑜, 宋军生. 不同基因型亚麻下胚轴不定芽诱导的研究, 作物杂志, 2014, (3):39−43.Chen F, Dang Z H, Zhang J P, Li W J, Hao R K, Zhang Q, Zhang Y, Song J S. Studies on the induction of adventitious shoots in hypocotyls of flax from different genotypes. Crops, 2014, (3): 39−43 (in Chinese with English abstract).
[59] 李闻娟, 齐燕妮, 王利民, 党照, 赵利, 赵玮, 谢亚萍, 王斌, 张建平, 李淑洁. 不同胡麻品种 TAG 合成途径关键基因表达与含油量、脂肪酸组分的相关性分析. 草业学报, 2019, 28(1): 138−149.Li W J, Qi Y N, Wang L M, Dang Z, Zhao L, Zhao W, Xie Y P, Wang B, Zhang J P, Li S J. Correlation analysis between the expression of key genes of TAG synthesis pathway and oil content and fatty acid fractions in different caraway varieties. Acta Pratac Sin, 2019. 28(1): 138-149 (in Chinese with English abstract).
[60] 范鑫, 赵雷霖, 翟红红, 王远, 孟志刚, 梁成真, 张锐, 郭三堆, 孙国清. AtNEK6 在棉花旱盐胁迫响应中的表达分析研究. 中国农业科学, 2018, 51: 4230−4240.Fan X, Zhao L L, Zhai H H, Wang Y, Meng Z G, Liang C Z, Zhang R, Guo S D, SUN G Q. Study on expression analysis of AtNEK6 in response to drought and salt stress in cotton. Scientia Aguicultura Sinica, 2018, 51: 4230−4240 (in Chinese with English abstract).
[61] Fahad S, Bajwa A A, Nazir U, Anjum S A, Farooq A, Zohaib A, Sadia S, Nasim W, Adkins S, Saud S, Ihsan M Z, Alharby H, Wu C, Wang D, Huang J. Crop production under drought and heat stress: plant responses and management options. Front Plant Sci, 2017, 8: 1147.
[62] Cernac A, Benning C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J, 2004, 40: 575−585.
[63] Shen B, Allen W B, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski M C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol, 2010, 153: 980−987.
[64] Liu J, Hua W, Zhan G, Wei F, Wang X, Liu G, Wang H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Biochem, 2010, 48: 9−15.
[65] 邵宇鹏, 杨明明, 包格格, 孙英楠, 杨强, 李文滨, 王志坤. 大豆GmWRI1a 基因启动子克隆及其功能分析. 中国油料作物学报, 2019, 41:517−523.Shao Y P, Yang M M, Bao G G, Sun Y N, Yang Q, Li W B, Wang Z K. Cloning of soybean GmWRI1a gene promoter and its functional analysis.Chinese Journal of Oil Crop Sciences, 2019, 41: 517−523 (in Chinese with English abstract).
[66] 闫丽, 杨强, 邵宇鹏, 李丹丹, 王志坤, 李文滨. 大豆 GmWRI1a 基因启动子克隆及序列分析. 作物杂志, 2017, (2): 51−58.Yan L, Yang Q, Shao Y P, Li D D, Wang Z K, Li W B. Cloning and sequence analysis of soybean GmWRI1a gene promoter. Crops, 2017, (2): 51−58 (in Chinese with English abstract).
[67] 李丹丹, 闫丽, 常健敏, 王志坤, 李文滨. 大豆GmWRI1 基因在糖,植物激素及盐胁迫下的表达分析. 作物杂志, 2015, (4): 41−46.Li D D, Yan L, Chang J M, Wang Z K, Li W B. Expression analysis of soybean GmWRI1 gene under sugar,phytohormone and salt stress. Crops, 2015, (4): 41−46 (in Chinese with English abstract).
[67] 郝翠翠, 花生转录因子 AhWRI1 基因的克隆与功能研究. 青岛科技大学硕士学位论文, 山东青岛, 2018.Hao C C. Cloning and Functional Study of Peanut Transcription Factor AhWRI1 Gene. MS Thesis of Qingdao University of Science and Technology, Qingdao, Shangdong, China, 2018 (in Chinese with English abstract).
[69] Arias-Moreno D M, Jiménez-Bremont J F, Maruri-López I, Delgado-Sánchez P. Effects of catalase on chloroplast arrangement in Opuntia streptacantha chlorenchyma cells under salt stress. Sci Rep, 2017, 7: 8656.
[70] Choudhury F K, Rivero R M, Blumwald E, Mittler R. Reactive oxygen species, abiotic stress and stress combination. Plant J, 2017, 90: 856−867.
[71] Xing X, Zhou Q, Xing H, Jiang H, Wang S. Early abscisic acid accumulation regulates ascorbate and glutathione metabolism in soybean leaves under progressive water stress. J Plant Growth Regul, 2016, 35: 865−876.
[72] 牟舒敏, 张丽娟, 李红兵, 关月明, 可庆波, 张岁岐, 郭尚洙, 邓西平. 三种转基因甘薯响应PEG-6000 模拟干旱胁迫的生理性差异. 植物生理学报, 2023, 59: 1339−1350.Mou S M, Zhang L J, Li H B, Guan Y M, Ke Q B, Zhang Y Q, Guo S S, Deng X P. Physiological differences among three transgenic sweetpotatoes in response to PEG-6000-mimicked drought stress. J Plant Physiol, 2023, 59: 1339−1350 (in Chinese with English abstract).
文章摘自: 李闻娟,王利民,齐燕妮,赵玮,谢亚萍,党照,赵丽蓉,李雯,徐晨梦,王琰,张建平.亚麻LuWRI1a在旱盐胁迫响应中的功能分析[J/OL].作物学报.1-14[2024-02-20]. https://link.cnki.net/urlid/11.1809.s.20240219.1923.028